Lighthouse LabsW7D5 - Pipelines and Model PersistenceInstructor: Socorro E. Dominguez-Vidana |

|
Overview - Pipelines¶
- [] Motivation and example
- [] Feature unions
- [] Column transformers
- [] Visualizing pipelines
- [] Hyperparameter tuning with pipelines
- [] Custom class in a pipeline

| Mario and Luigi have been hired to get water flowing smoothly through a set of pipes. Each part of the job has to be done in the right order, and they use their plumbing pipeline to keep it organized. |
Clear the Pipes Luigi goes first, clearing out any clogs and making sure the pipes are clean. In a ML pipeline, this is like cleaning the data
Check the Pipe Sizes Next, Mario measures and standardizes the pipes to ensure they're compatible. This step would be similar to scaling or normalizing the data.
Install Filters To keep the water clean, Luigi adds filters at specific points in the pipeline. Similarly, we may add steps to transform data or select only the features we need, so only relevant information goes through.
Test the Water Flow Before they finish, Mario and Luigi test the water flow to make sure everything works. In ML, this step is like training and testing a model guaranteeing that our setup can handle real-world data.
Final Check and Save the Setup Finally, they document how they set up the pipes so anyone can understand it if they need repairs later. In ML, we can save the pipeline model with
jobliborpickleso we can use it again without setting it up from scratch.
Why Use a Pipeline?¶
- Reuse steps easily for new data.
- Test everything consistently and know the exact order, so there are no surprises in model performance.
In simple terms, an ML pipeline is our plumbing plan for handling data from start to finish, making sure every step flows correctly.
Using pipelines¶
Besides being a plumber, Mario took up another role: Doctor Mario!
The famous plumber-turned-physician is on a new mission: tackling diabetes! In his clinic, he got access to the diabetes dataset that holds information on patients' age, blood pressure, BMI, and more. With this data, Dr. Mario can identify risk factors, predict who might be at risk, and create better treatment plans.
Just like fixing a tricky pipe, understanding diabetes requires the right tools and steps.
import pandas as pd
df = pd.read_csv('data/diabetes.csv')
df.head()
| preg | plas | pres | skin | test | mass | pedi | age | class | |
|---|---|---|---|---|---|---|---|---|---|
| 0 | 6 | 148 | 72 | 35 | 0 | 33.6 | 0.627 | 50 | 1 |
| 1 | 1 | 85 | 66 | 29 | 0 | 26.6 | 0.351 | 31 | 0 |
| 2 | 8 | 183 | 64 | 0 | 0 | 23.3 | 0.672 | 32 | 1 |
| 3 | 1 | 89 | 66 | 23 | 94 | 28.1 | 0.167 | 21 | 0 |
| 4 | 0 | 137 | 40 | 35 | 168 | 43.1 | 2.288 | 33 | 1 |
from sklearn.model_selection import train_test_split
from sklearn.preprocessing import StandardScaler
from sklearn.decomposition import PCA
from sklearn.linear_model import LogisticRegression, RidgeClassifier
from sklearn.metrics import accuracy_score
Without a Pipeline¶
X = df.drop(columns='class')
y = df['class']
X_train, X_test, y_train, y_test = train_test_split(X, y, test_size=0.2, random_state=27, stratify=y)
scaler = StandardScaler()
scaler.fit(X_train)
X_train_scaled = scaler.transform(X_train)
pca = PCA(n_components=3)
pca.fit(X_train_scaled)
X_train_pca = pca.transform(X_train_scaled)
model = LogisticRegression()
model.fit(X_train_pca, y_train)
# Test portion
X_test_scaled = scaler.transform(X_test)
X_test_pca = pca.transform(X_test_scaled)
y_pred = model.predict(X_test_pca)
acc = accuracy_score(y_test, y_pred)
print(f'Test set accuracy: {acc}')
Test set accuracy: 0.6948051948051948
There are several inconvenient things about this: then our model will not work as expected.
- We have a lot of ugly code. We keep calling
.fit()and.transform()on different objects, and we keep having to rename transformed variables so as not to cause confusions later in our notebook. - Our preprocessing and modeling code is distributed and therefore error-prone. If we try running our model somewhere else and forget to copy over a step (e.g. we don't apply StandardScaler to the test set),
- We can only use convenient Sklearn functions/classes such as
GridSearchCV()on the model class (e.g. LogisticRegression). What if we want to try different numbers of components, or different scaling methods?
The solution: Sklearn Pipelines¶
from sklearn.pipeline import Pipeline
pipeline = Pipeline(steps=[('scaling', StandardScaler()),
('pca', PCA(n_components=3)),
('classifier', LogisticRegression())])
pipeline.fit(X_train, y_train)
y_pred = pipeline.predict(X_test)
acc = accuracy_score(y_test, y_pred)
print(f'Test set accuracy: {acc}')
Test set accuracy: 0.6948051948051948
Notice how much cleaner this code is. The composite model created using Pipeline
can be used just like any other Sklearn model you have learned, which means that it
can also be passed to functions like cross_val_score().
Feature unions¶
Pipeline lets us specify a sequence of steps that will be executed in one after the other (i.e. in series). But it wouldn't make sense if Mario always had to wait for Luigi to finish a job. Sometimes, Mario and Luigi can each work on different parts of a job at the same time, then combining their results at the end.
In ML, sometimes we want to apply different transformations to our data in parallel and then combine the results. We might want to scale numerical features (like age or pregnancies) while applying one-hot encoding to categorical features (like gender or city). Instead of doing this in separate steps, FeatureUnion lets us do both at once and then joins them together.
FeatureUnion's can be composed with Pipeline's however much we want.
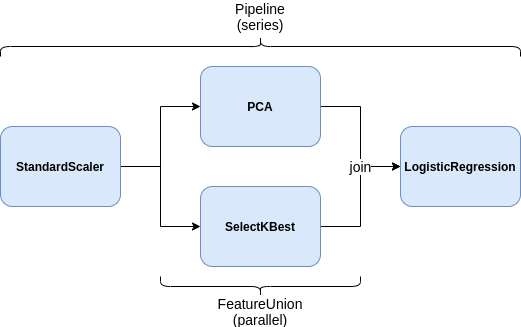
from sklearn.pipeline import FeatureUnion
from sklearn.feature_selection import SelectKBest
feature_union = FeatureUnion([('pca', PCA(n_components=3)),
('select_best', SelectKBest(k=6))])
pipeline = Pipeline(steps=[('scaling', StandardScaler()),
('features', feature_union),
('classifier', LogisticRegression())])
pipeline.fit(X_train, y_train)
y_pred = pipeline.predict(X_test)
acc = accuracy_score(y_test, y_pred)
print(f'Test set accuracy: {acc}')
Test set accuracy: 0.7337662337662337
Column transformers¶
FeatureUnion doesn’t automatically "split" the data between transformers. It applies each transformer to the full dataset. For different transformations applied to specific parts of the data, we use ColumnTransformer, which allows you to specify which columns should go to which transformation.
For instance, for a dataset with both numerical and categorical features, we may want to do something like the following:
- For numeric columns:
- Impute missing values with the mean
- Standard scale the values
- For categorical columns:
- Impute missing values with the mode
- One-hot-encode the categories
- Fit a model to the resulting features
ColumnTransformer has a very similar syntax to that of a FeatureUnion or Pipeline, except that we must also specify the column names that each transform applies to.
Doctor Mario is tackling heart disease now! With patient data on age, cholesterol, heart rate, and more, he aims to identify key risk factors to help his patients.
| Feature | Description |
|---|---|
age |
Age of the patient in years |
sex |
Sex of the patient (1 = male, 0 = female) |
cp |
Chest pain type (0: typical angina, 1: atypical angina, 2: non-anginal pain, 3: asymptomatic) |
trestbps |
Resting blood pressure in mm Hg |
chol |
Serum cholesterol in mg/dl |
fbs |
Fasting blood sugar > 120 mg/dl (1 = true, 0 = false) |
restecg |
Resting electrocardiographic results (0: normal, 1: ST-T wave abnormality, 2: probable left ventricular hypertrophy) |
thalach |
Maximum heart rate achieved |
exang |
Exercise-induced angina (1 = yes, 0 = no) |
oldpeak |
ST depression induced by exercise relative to rest |
slope |
Slope of the peak exercise ST segment (0: upsloping, 1: flat, 2: downsloping) |
ca |
Number of major vessels (0-3) colored by fluoroscopy |
thal |
Thalassemia (3 = normal, 6 = fixed defect, 7 = reversible defect) |
target |
Diagnosis of heart disease (1 = disease, 0 = no disease) |
df = pd.read_csv('data/heart_disease.csv')
df.head()
| age | sex | cp | trestbps | chol | fbs | restecg | thalach | exang | oldpeak | slope | ca | thal | target | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63.0 | 1.0 | 1.0 | 145.0 | 233.0 | 1.0 | 2.0 | 150.0 | 0.0 | 2.3 | 3.0 | 0.0 | 6.0 | 0 |
| 1 | 67.0 | 1.0 | 4.0 | 160.0 | 286.0 | 0.0 | 2.0 | 108.0 | 1.0 | 1.5 | 2.0 | 3.0 | 3.0 | 2 |
| 2 | 67.0 | 1.0 | 4.0 | 120.0 | 229.0 | 0.0 | 2.0 | 129.0 | 1.0 | 2.6 | 2.0 | 2.0 | 7.0 | 1 |
| 3 | 37.0 | 1.0 | 3.0 | 130.0 | 250.0 | 0.0 | 0.0 | 187.0 | 0.0 | 3.5 | 3.0 | 0.0 | 3.0 | 0 |
| 4 | 41.0 | 0.0 | 2.0 | 130.0 | 204.0 | 0.0 | 2.0 | 172.0 | 0.0 | 1.4 | 1.0 | 0.0 | 3.0 | 0 |
X, y = df.drop(columns='target'), df['target'].values
X_train, X_test, y_train, y_test = train_test_split(X, y, random_state=27)
from sklearn.compose import ColumnTransformer
from sklearn.preprocessing import OneHotEncoder
from sklearn.impute import SimpleImputer
numerical_features = ["age", "trestbps", "chol", "thalach", "oldpeak"]
categorical_features = ["sex", "cp", "fbs", "restecg", "exang", "slope", "ca", "thal"]
numeric_transform = StandardScaler()
categorical_transform = OneHotEncoder(handle_unknown="ignore")
preprocessor = ColumnTransformer([('numeric', numeric_transform, numerical_features),
('categorical', categorical_transform, categorical_features)])
pipeline = Pipeline(steps=[
("preprocessor", preprocessor),
("classifier", LogisticRegression())
])
pipeline.fit(X_train, y_train)
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol',
'thalach', 'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs',
'restecg', 'exang', 'slope',
'ca', 'thal'])])),
('classifier', LogisticRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol',
'thalach', 'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs',
'restecg', 'exang', 'slope',
'ca', 'thal'])])),
('classifier', LogisticRegression())])ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol', 'thalach',
'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs', 'restecg', 'exang',
'slope', 'ca', 'thal'])])['age', 'trestbps', 'chol', 'thalach', 'oldpeak']
StandardScaler()
['sex', 'cp', 'fbs', 'restecg', 'exang', 'slope', 'ca', 'thal']
OneHotEncoder(handle_unknown='ignore')
LogisticRegression()
What is the ColumnTransformer actually doing? We can get a better sense by looking at our data before and after
the transform it applies.
from sklearn.metrics import accuracy_score
y_pred = pipeline.predict(X_test)
acc = accuracy_score(y_test, y_pred)
print(f"Test set accuracy: {acc}")
Test set accuracy: 0.618421052631579
# Initial data
X.head()
| age | sex | cp | trestbps | chol | fbs | restecg | thalach | exang | oldpeak | slope | ca | thal | |
|---|---|---|---|---|---|---|---|---|---|---|---|---|---|
| 0 | 63.0 | 1.0 | 1.0 | 145.0 | 233.0 | 1.0 | 2.0 | 150.0 | 0.0 | 2.3 | 3.0 | 0.0 | 6.0 |
| 1 | 67.0 | 1.0 | 4.0 | 160.0 | 286.0 | 0.0 | 2.0 | 108.0 | 1.0 | 1.5 | 2.0 | 3.0 | 3.0 |
| 2 | 67.0 | 1.0 | 4.0 | 120.0 | 229.0 | 0.0 | 2.0 | 129.0 | 1.0 | 2.6 | 2.0 | 2.0 | 7.0 |
| 3 | 37.0 | 1.0 | 3.0 | 130.0 | 250.0 | 0.0 | 0.0 | 187.0 | 0.0 | 3.5 | 3.0 | 0.0 | 3.0 |
| 4 | 41.0 | 0.0 | 2.0 | 130.0 | 204.0 | 0.0 | 2.0 | 172.0 | 0.0 | 1.4 | 1.0 | 0.0 | 3.0 |
# Preprocessed data
X_preprocessed = preprocessor.transform(X)
X_preprocessed[0]
array([ 0.92303809, 0.78129791, -0.2614227 , 0.08134766, 1.07875303,
0. , 1. , 1. , 0. , 0. ,
0. , 0. , 1. , 0. , 0. ,
1. , 1. , 0. , 0. , 0. ,
1. , 1. , 0. , 0. , 0. ,
0. , 0. , 1. , 0. , 0. ])
Visualizing pipelines¶
Another advantage of having these pipelines is that we can quickly visualize complex workflows used in our modeling as HTML, which can be helpful for debugging purposes or presentations.
Note: I highly recommend you use this in your own presentations as a substitute for (or in addition to) code.
# Display HTML representation in a jupyter context
from sklearn import set_config
set_config(display='diagram')
pipeline
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol',
'thalach', 'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs',
'restecg', 'exang', 'slope',
'ca', 'thal'])])),
('classifier', LogisticRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('preprocessor',
ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol',
'thalach', 'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs',
'restecg', 'exang', 'slope',
'ca', 'thal'])])),
('classifier', LogisticRegression())])ColumnTransformer(transformers=[('numeric', StandardScaler(),
['age', 'trestbps', 'chol', 'thalach',
'oldpeak']),
('categorical',
OneHotEncoder(handle_unknown='ignore'),
['sex', 'cp', 'fbs', 'restecg', 'exang',
'slope', 'ca', 'thal'])])['age', 'trestbps', 'chol', 'thalach', 'oldpeak']
StandardScaler()
['sex', 'cp', 'fbs', 'restecg', 'exang', 'slope', 'ca', 'thal']
OneHotEncoder(handle_unknown='ignore')
LogisticRegression()
Note that you can also click on the individual parts in the diagram (e.g. StandardScaler) to see their arguments.
# Or, save the HTML to a file
from sklearn.utils import estimator_html_repr
with open('img/model_pipeline.html', 'w') as f:
f.write(estimator_html_repr(pipeline))
Hyperparameter tuning with pipelines¶

|
Normally, if we want to tune hyperparameters using something like
When not using pipelines, we can only tune hyperparameters for a single model (the one we specify as the
model in |
from sklearn.model_selection import GridSearchCV, cross_val_score
pipeline = Pipeline(steps=[
('imputer', SimpleImputer(strategy='mean')),
('scaling', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression())
])
# Define the parameter grid
param_grid = {
'pca__n_components': [2, 3, 4],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs']}
# GridSearchCV with cross-validation
grid_search = GridSearchCV(estimator=pipeline, param_grid=param_grid, cv=5, scoring='accuracy')
grid_search.fit(X_train, y_train)
print(f'Best Parameters: {grid_search.best_params_}')
print(f'Best Cross-Validation Score: {grid_search.best_score_}')
# Evaluate the test set with the best model found
best_pipeline = grid_search.best_estimator_
acc = best_pipeline.score(X_test, y_test)
print(f'Test set accuracy with best parameters: {acc}')
Best Parameters: {'classifier__C': 0.1, 'classifier__solver': 'liblinear', 'pca__n_components': 3}
Best Cross-Validation Score: 0.6125603864734299
Test set accuracy with best parameters: 0.5921052631578947
In addition to trying out different hyperparameters for a given step in the pipeline, you can also try different classes altogether. For instance, what if we wanted to try both Logistic Regression and SVM for the "classifier" step?
from sklearn.svm import SVC
pipeline = Pipeline([
('imputer', SimpleImputer(strategy='mean')),
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression()) # Placeholder for classifier
])
param_grid = [
{
'pca__n_components': [2, 3, 4],
'classifier': [LogisticRegression()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs']
},
{
'pca__n_components': [2, 3, 4],
'classifier': [SVC()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__kernel': ['linear', 'rbf']
},
{
'pca__n_components': [2, 3, 4],
'classifier': [RidgeClassifier()],
'classifier__alpha': [0.1, 0.01, 1.0]
}
]
grid_search = GridSearchCV(pipeline, param_grid, cv=5, scoring='accuracy')
grid_search.fit(X_train, y_train)
GridSearchCV(cv=5,
estimator=Pipeline(steps=[('imputer', SimpleImputer()),
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression())]),
param_grid=[{'classifier': [LogisticRegression()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs'],
'pca__n_components': [2, 3, 4]},
{'classifier': [SVC()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__kernel': ['linear', 'rbf'],
'pca__n_components': [2, 3, 4]},
{'classifier': [RidgeClassifier()],
'classifier__alpha': [0.1, 0.01, 1.0],
'pca__n_components': [2, 3, 4]}],
scoring='accuracy')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=5,
estimator=Pipeline(steps=[('imputer', SimpleImputer()),
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression())]),
param_grid=[{'classifier': [LogisticRegression()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs'],
'pca__n_components': [2, 3, 4]},
{'classifier': [SVC()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__kernel': ['linear', 'rbf'],
'pca__n_components': [2, 3, 4]},
{'classifier': [RidgeClassifier()],
'classifier__alpha': [0.1, 0.01, 1.0],
'pca__n_components': [2, 3, 4]}],
scoring='accuracy')Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler()),
('pca', PCA(n_components=2)), ('classifier', SVC(C=10))])SimpleImputer()
StandardScaler()
PCA(n_components=2)
SVC(C=10)
print("Best Parameters:", grid_search.best_params_)
print("Best Cross-Validation Score:", grid_search.best_score_)
Best Parameters: {'classifier': SVC(), 'classifier__C': 10, 'classifier__kernel': 'rbf', 'pca__n_components': 2}
Best Cross-Validation Score: 0.6302415458937198
y_pred = grid_search.best_estimator_.predict(X_test)
acc = accuracy_score(y_test, y_pred)
print(f"Test set accuracy: {acc}")
Test set accuracy: 0.5921052631578947
Model Persistence¶
After hours of fixing pipes, Mario and Luigi don't want to start from scratch if they have to revisit the job. In ML, pipelines can take a long time to .fit, and you will not want to run the whole notebook every single time - we want to save the model so we can use it again without retraining.
This process is called model persistence. Just like Mario and Luigi might record their work notes, we use pickle or joblib to save models. This lets us load them back up later ready to make predictions.
Serialization is the process of converting a program entity into a stream of bytes that can be saved as a file. Serialization:
- Avoids redundant training. Models can take a long time to train and data can take a long time to load/process
- allows us to deploy the model into an application.
Pickle¶
- Pickling is the process where a Python object is converted into a byte stream (usually not human readable).
- Unpickling is the reverse operation, where a byte stream is converted back into a working Python object.
- Pickling is the simplest way to store the object from a coding perspective.
- The Python Pickle module is an object-oriented way of storing objects.
- It can store any Python object, not just
Sklearnmodels.
- It can store any Python object, not just
Features¶
- Store/load dictionaries and lists.
- Store/load the attributes of arbitrary data types (i.e. classes)
- Do this recursively, so that if your object has attributes that are classes themselves, it can be saved just as easily
Limitations¶
- Does not save the code of an object — only its attribute values.
- Cannot save file handles or connection sockets.
- Pickle is version-dependent. For example, if you saved a model with a certain version of
Sklearnthen try to load it with a different one (e.g. you updated), there may be issues.- Another motivation for using virtual environments, which can be containerized.
Saving procedure¶
import pickle # Built-in python module
# Create some object and manipulate it in some way (e.g. train the model)
myobj = SomeClass(...)
myobj = myobj.some_method(...)
# Save to a file using Pickle
with open('myfile.pickle', 'wb') as file_handle:
pickle.dump(myobj, file_handle)
Loading procedure¶
import pickle # Built-in python module
# Load from a file using Pickle
with open('myfile.pickle', 'rb') as file_handle:
myobj = pickle.load(file_handle) # myobj will be an instance of SomeClass
Methods¶
The pickle module provides four different methods:
- dump() − The dump() method serializes to an open file (file-like object).
- dumps() − Serializes to a string.
- load() − Deserializes from an open-like object.
- loads() − Deserializes from a string.
Example¶
import pickle
# Save the model
with open('saved_models/pipeline.pickle', 'wb') as f:
pickle.dump(pipeline, f)
# Load the model
with open('saved_models/pipeline.pickle', 'rb') as f:
pipeline_loaded = pickle.load(f)
pipeline_loaded
Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler()),
('pca', PCA()), ('classifier', LogisticRegression())])In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler()),
('pca', PCA()), ('classifier', LogisticRegression())])SimpleImputer()
StandardScaler()
PCA()
LogisticRegression()
Joblib¶
Joblib is an alternative serialization module to Pickle. However, starting with Python 3.8, Pickle is actually better than Joblib for saving numpy arrays.
If you have Python >=3.8, just use Pickle. Source.*
Saving procedure¶
import joblib
# Create some object and manipulate it in some way (e.g. train the model)
myobj = SomeClass(...)
myobj = myobj.some_method(...)
# Save to a file using Joblib
joblib.dump(myobj, file_path)
Loading procedure¶
import joblib
# Load from a file using Joblib
myobj = joblib.load(file_path) # myobj will be an instance of SomeClass
Example¶
import joblib
joblib.dump(grid_search, 'saved_models/pipeline.can')
# Load the model
pipeline_loaded = joblib.load('saved_models/pipeline.can')
pipeline_loaded
GridSearchCV(cv=5,
estimator=Pipeline(steps=[('imputer', SimpleImputer()),
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression())]),
param_grid=[{'classifier': [LogisticRegression()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs'],
'pca__n_components': [2, 3, 4]},
{'classifier': [SVC()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__kernel': ['linear', 'rbf'],
'pca__n_components': [2, 3, 4]},
{'classifier': [RidgeClassifier()],
'classifier__alpha': [0.1, 0.01, 1.0],
'pca__n_components': [2, 3, 4]}],
scoring='accuracy')In a Jupyter environment, please rerun this cell to show the HTML representation or trust the notebook. On GitHub, the HTML representation is unable to render, please try loading this page with nbviewer.org.
GridSearchCV(cv=5,
estimator=Pipeline(steps=[('imputer', SimpleImputer()),
('scaler', StandardScaler()),
('pca', PCA()),
('classifier', LogisticRegression())]),
param_grid=[{'classifier': [LogisticRegression()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__solver': ['liblinear', 'lbfgs'],
'pca__n_components': [2, 3, 4]},
{'classifier': [SVC()],
'classifier__C': [0.1, 1, 10, 100],
'classifier__kernel': ['linear', 'rbf'],
'pca__n_components': [2, 3, 4]},
{'classifier': [RidgeClassifier()],
'classifier__alpha': [0.1, 0.01, 1.0],
'pca__n_components': [2, 3, 4]}],
scoring='accuracy')Pipeline(steps=[('imputer', SimpleImputer()), ('scaler', StandardScaler()),
('pca', PCA(n_components=2)), ('classifier', SVC(C=10))])SimpleImputer()
StandardScaler()
PCA(n_components=2)
SVC(C=10)